Qian Y, Arellano G, Ifergan I, Lin J, Snowden C, Kim T, Thomas JJ, Law C, Guan T, Balabanov RD, Kaech SM, Miller SD, Choi J. ZEB1 promotes pathogenic Th1 and Th17 cell differentiation in multiple sclerosis. Cell Rep. 2021 Aug 24;36(8):109602. doi: 10.1016/j.celrep.2021.109602. PMID: 34433042; PMCID: PMC8431781.
Note: Acknowledges Admera Health “for their invaluable contributions”
Andersen J, Revah O, Miura Y, Thom N, Amin ND, Kelley KW, Singh M, Chen X, Thete MV, Walczak EM, Vogel H, Fan HC, Paşca SP. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell. 2020 Dec 23;183(7):1913-1929.e26. doi: 10.1016/j.cell.2020.11.017. Epub 2020 Dec 16. PMID: 33333020; PMCID: PMC8711252.
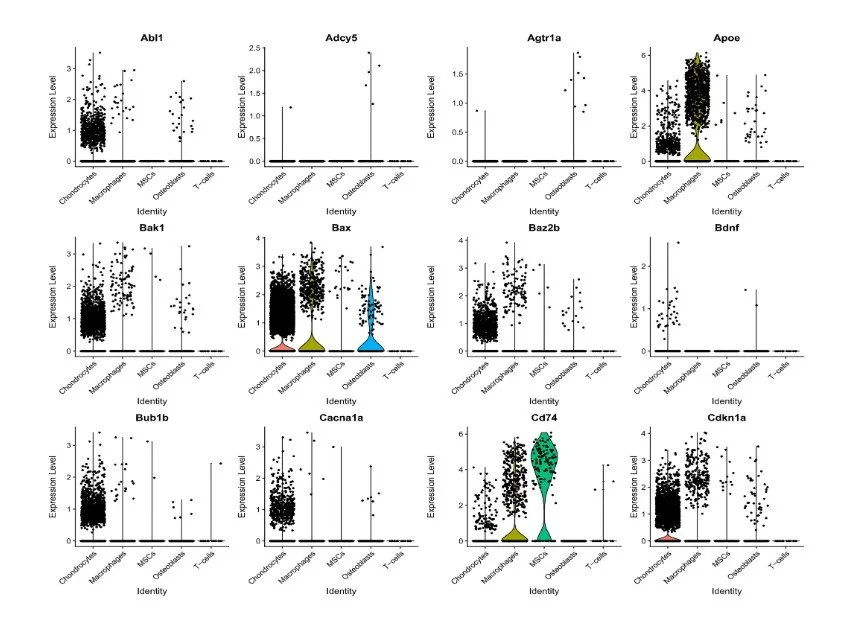
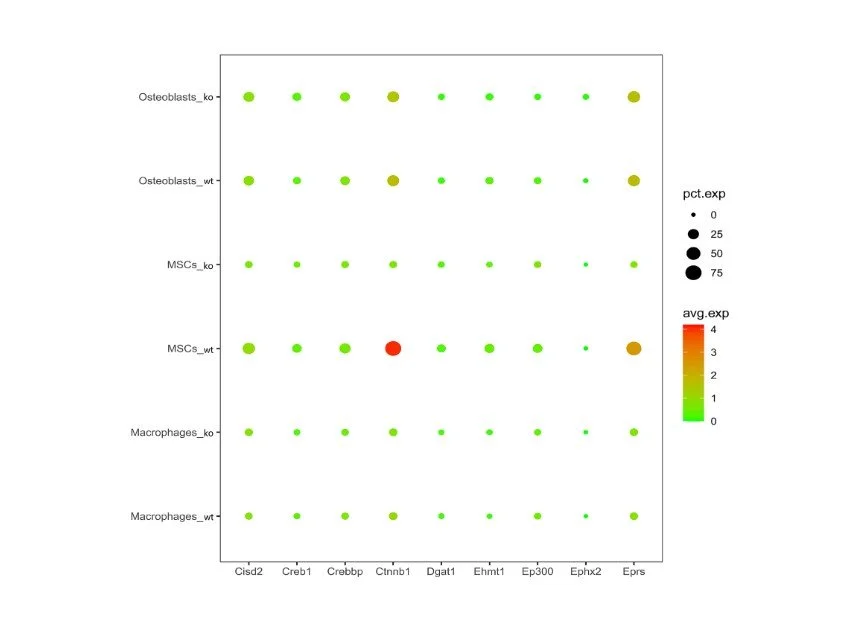
Dissociated cells were resuspended in ice-cold PBS containing 0.02% BSA and loaded onto a Chromium Single cell 3′ chip (with an estimated recovery of 6,000 cells per channel) to generate gel beads in emulsion (GEMs). scRNA-seq libraires were prepared with the Chromium Single cell 3′ GEM, Library & Gel Bead Kit v3 (10x Genomics, PN: 1000075). Libraries from different samples were pooled and sequenced by Admera Health on a NovaSeq S4 (Illumina) using 150 × 2 chemistry. Demultiplexing, alignment, barcode and UMI counting and aggregation were performed using Cell Ranger (v3.0 with default settings). This provided us with a feature by count matrix.
Khan TA, Revah O, Gordon A, Yoon SJ, Krawisz AK, Goold C, Sun Y, Kim CH, Tian Y, Li MY, Schaepe JM, Ikeda K, Amin ND, Sakai N, Yazawa M, Kushan L, Nishino S, Porteus MH, Rapoport JL, Bernstein JA, O'Hara R, Bearden CE, Hallmayer JF, Huguenard JR, Geschwind DH, Dolmetsch RE, Paşca SP. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat Med. 2020 Dec;26(12):1888-1898. doi: 10.1038/s41591-020-1043-9. Epub 2020 Sep 28. PMID: 32989314; PMCID: PMC8525897.
Bead Kit v2(10x Genomics, PN:120237). Libraries from different samples were pooled and sequenced by Admera Health on a NovaSeq S4 (Illumina) using 150*2 chemistry.
Revah O, Gore F, Kelley KW, Andersen J, Sakai N, Chen X, Li MY, Birey F, Yang X, Saw NL, Baker SW, Amin ND, Kulkarni S, Mudipalli R, Cui B, Nishino S, Grant GA, Knowles JK, Shamloo M, Huguenard JR, Deisseroth K, Pașca SP. Maturation and circuit integration of transplanted human cortical organoids. Nature. 2022 Oct;610(7931):319-326. doi: 10.1038/s41586-022-05277-w. Epub 2022 Oct 12. PMID: 36224417; PMCID: PMC9556304.
snRNA-seq libraries were prepared with the Chromium Single cell 3′ GEM, Library & Gel Bead Kit v3 (10x Genomics). Libraries from different samples were pooled and sequenced by Admera Health on a NovaSeq S4 (Illumina).
Castelli M, Sabaneyeva E, Lanzoni O, Lebedeva N, Floriano AM, Gaiarsa S, Benken K, Modeo L, Bandi C, Potekhin A, Sassera D, Petroni G. Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales. ISME J. 2019 Sep;13(9):2280-2294. doi: 10.1038/s41396-019-0433-9. Epub 2019 May 9. PMID: 31073215; PMCID: PMC6776064.
In order to reach sufficient DNA quantity, the extract was subjected to whole-genome amplification (WGA) with the REPLI-g Single Cell Kit (Qiagen). The WGA product was processed through a Nextera XT library and sequenced on an Illumina HiSeq X machine by Admera Health (South Plainfield, NJ, USA), producing 37,100,964 pairs of 150 bp reads.
Ma L, Liang C, Wang J, Chang Q, Wang Y, Zhang W, Du Y, Sadan J, Chen JF. Reversing lysosome-ribosome circuit dysregulation mitigates C9FTD/ALS neurodegeneration and behaviors. Hum Mol Genet. 2023 Apr 6;32(8):1252-1265. doi: 10.1093/hmg/ddac271. PMID: 36322143; PMCID: PMC10077508.
Andersen J, Thom N, Shadrach JL, Chen X, Onesto MM, Amin ND, Yoon SJ, Li L, Greenleaf WJ, Müller F, Pașca AM, Kaltschmidt JA, Pașca SP. Single-cell transcriptomic landscape of the developing human spinal cord. Nat Neurosci. 2023 May;26(5):902-914. doi: 10.1038/s41593-023-01311-w. Epub 2023 Apr 24. PMID: 37095394.